The program we will be using to run neural simulations is Neuron. It was developed by Michael Hines of Yale University over many years and has emerged as a useful tool especially for working with compartmental models of single neurons. We have modified Neuron to incorporate various ligand receptor mechanisms and a 5 ion-channel mechanism (the classic Hodgkin-Huxley mechanism has 2 ion channels) that accurately simulates the generation of action potentials in the Tiger Salamander retinal ganglion cell. We call this program spike.
If you simply type spike at the terminal you will get the prompt oc> . All commands to the program could be entered at this prompt, but generally one uses what is called a hoc file to specify a simulation. There are three major categories that need to be specified in any neural simulation: 1. The geometry of the cell, 2. The membrane mechanisms, and 3. The stimulus.
The hoc file soma.hoc will be used for illustration. Here we will simulate a curent clamp experiment in a single compartment (a soma) where there is single synaptic input. The first lines of the file specify the geometry:
create soma
access soma
L = 25
diam = 25
nseg=1
We are creating an entity called soma and then accessing that entity. We give it both a length and diameter of 25 microns using one segment (compartment).
Next we will insert a passive membrane mechanism with a specific membrane conductance of
1/70000 Siemens /cm2 and a reversal potential of -65 mv. and an axial resistance of 110 ohm-cm :
insert pas
g_pas=1/70000
e_pas=-65
Ra=110
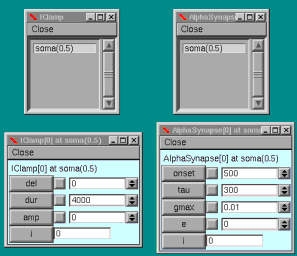
The next lines create a current clamp with no delay that continues for 4000 ms. using the IClamp point process . A point process is a Neuron entity that is placed at a specific point within a geometry; as opposed to a mechanism which like pas above is inserted at all "points" of the soma membrane. IClamp is placed in the middle of the soma by the (0.5) notation. Note the dot (.) notation used for specifying delay and duration. Amplitude also needs to be specified and is done so at a different level than the hoc file in this simulation.
objectvar clamp
clamp = new IClamp(0.5)
clamp.del = 0
clamp.dur = 4000
Finally the synapse is created using the AlphaSynapse point process in a manner similar to IClamp. The AlphaSynapse is the product of a linear and an exponential function to vary a conductance (g):
objectvar salpha
salpha = new AlphaSynapse(0.5)
salpha.onset = 500
salpha.tau = 300
salpha.gmax = 0.01
Now we can run the simulation by typing:
spike soma.hoc -
then at the prompt type:
oc> nrnmainmenu()

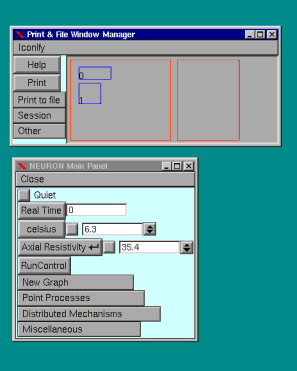
This will bring up the "Print & File Window Manager" and "Main Panel"
From this window various general functions can be executed.
This window allows you to open other windows needed to control and view the simulation.
The "celsius" and "Axial Resistivity" boxes are relevant only for Hodgkin-Huxley mechanisms.
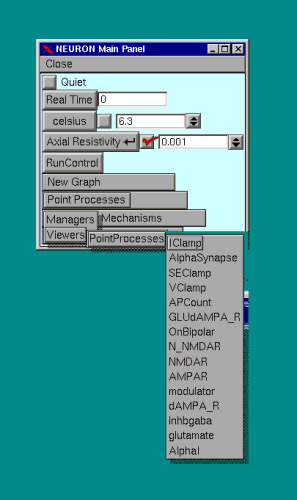
To view the point processes that have been inserted, choose the appropriate process from the "Main Panel" as shown
Under current clamp conditions we would like to see the variation in voltage. Under the Main Panel select New Graph / Voltage Axis to display a graphing window --
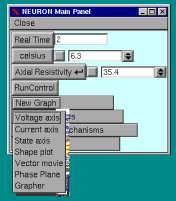
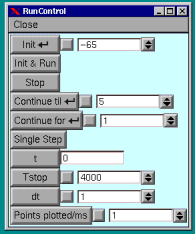
Selecting the RUN button on the Main Panel will bring up the Run Control menu.
Type in the run parameters as shown --
Init is the initial potential in mv.
Tstop is the run time of the simulation in ms.
dt is the integration step in ms.
Selecting Init & Run will start a single run of
4000 ms. duration. Doing this will demonstrate the voltage excursion caused by the Alpha Synapse without current clamp.
Note that we did not specifiy in the hoc file the amplitude of the current clamp. We could have included the line clamp.amp = 10 to give a 10 nA current, but generally we would like to give a family of clamping currents. We do this with the family function found under the Miscellaneous button of the Main Panel.
We will give 8 current steps ranging from
-0.08 to 0.2 nA
Selecting the Variable buuton allows you to choose clamp.amp as an independent variable.
Select Keep Lines in the graphing window and the Run in the family function control to begin the simulation.
To save the configuration of windows select Session / Save All from the "Print & File
Window Manager" and give the session the name soma.ses. The "session" will be brought-up when you execute "spike soma.hoc -" by adding this line to the end of the soma.hoc file:
xopen("soma.ses")
Further exercises will provide a session file.
There are two additional hoc files that will demonstrate the use of current clamp and voltage clamp. They are iclamp.hoc and vclamp.hoc respectively. Both of these are of similar form except for the type of clamp.
Both have a more complex geometry where a dendrite as well as a soma are defined. These two entities are connected by a line in the hoc file:
connect dend(0),soma(1)
Note that there are two Alpha Synapses defined. One in the soma and another in the dendrite.